MeTRN Help and Documentation
Contents
Download User ManualIntroduction
The cassava whole-genome transcriptional regulatory network (MeTRN) was built using three approaches: (1) template-based, (2) reverse engineering-based, and (3) cis-regulatory element-based methods. This database contains 4,812,519 transcriptional interactions between 33,006 protein-coding genes (covered 99.9% of all genes in the cassava genome) and 2,116 transcription factor genes. This database provide user to search on transcription regulators of interested target genes, target genes of transcription regulators, and MeTRN sub-network of interest pathways.
Tutorial
How to search for transcription regulators of target genes?
- Enter a single gene ID (based on genome version 6.1; Phytozome database)
- Press "Search" button
- Look search results
How to search for target genes of transcription factor?
- Select "Finding target gene from TF"
- Enter a single TF gene ID (based on genome version 6.1; Phytozome database)
- Press "Search" button
- Look search results
How to search for transcription regulators related to a pathway of interest?
- Select "Finding TF-target interaction related to a pathway"
- Type pathway name
- Press "Search" button
- Look search results
We divided the search results into two sessions: (i) gene information and (ii) TRN interaction information in table and network view.
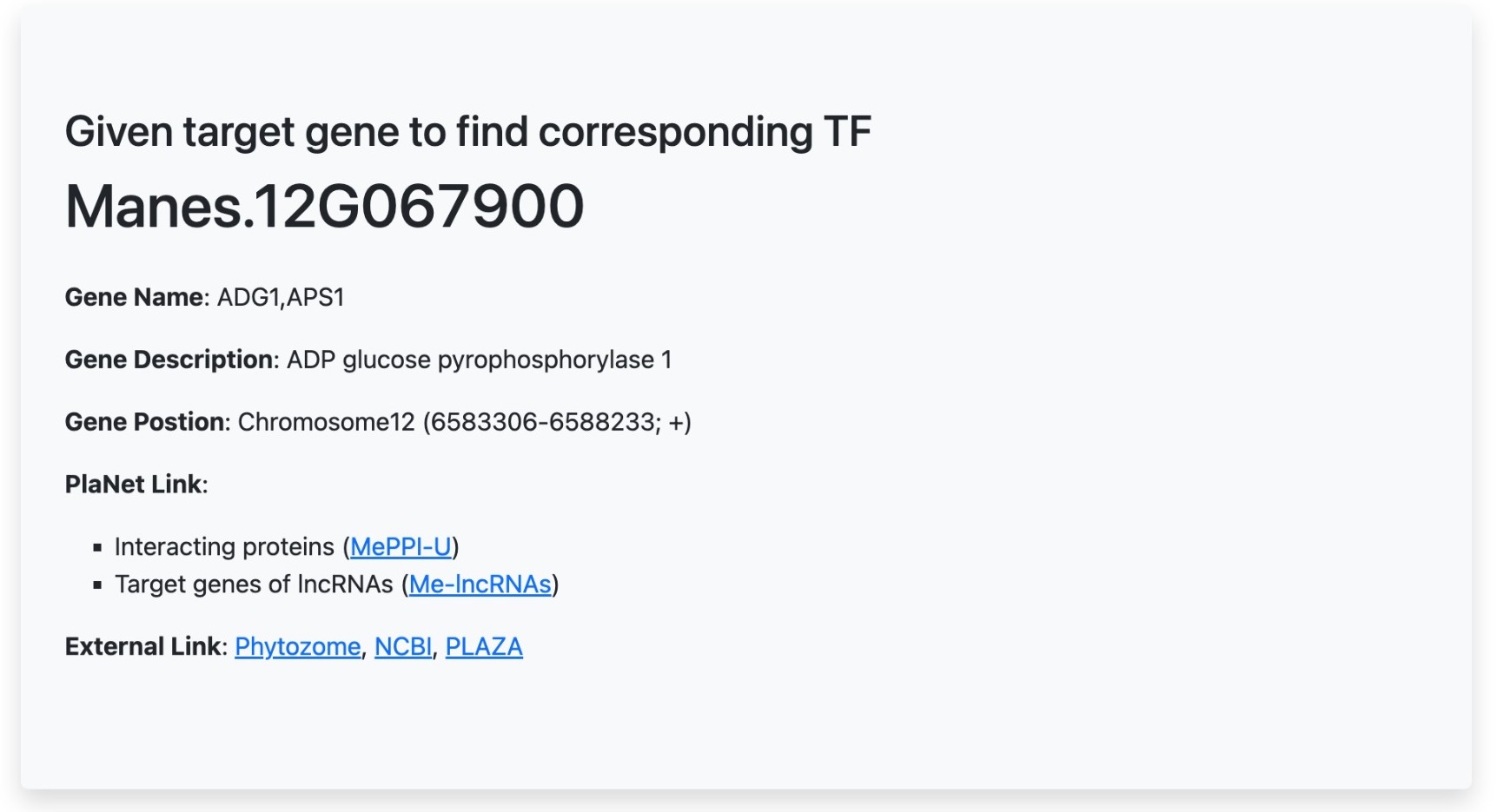
Gene information:
This session show input gene information, including,
- Gene name
- Gene description
- Transcription factor family (if that gene is transcription factor)
- Position of a gene in the cassava genome
- PlaNet link to MePPI-U, Me-LncRNAs, and MeRecon databases
- External link to Phytozome and NCBI databases
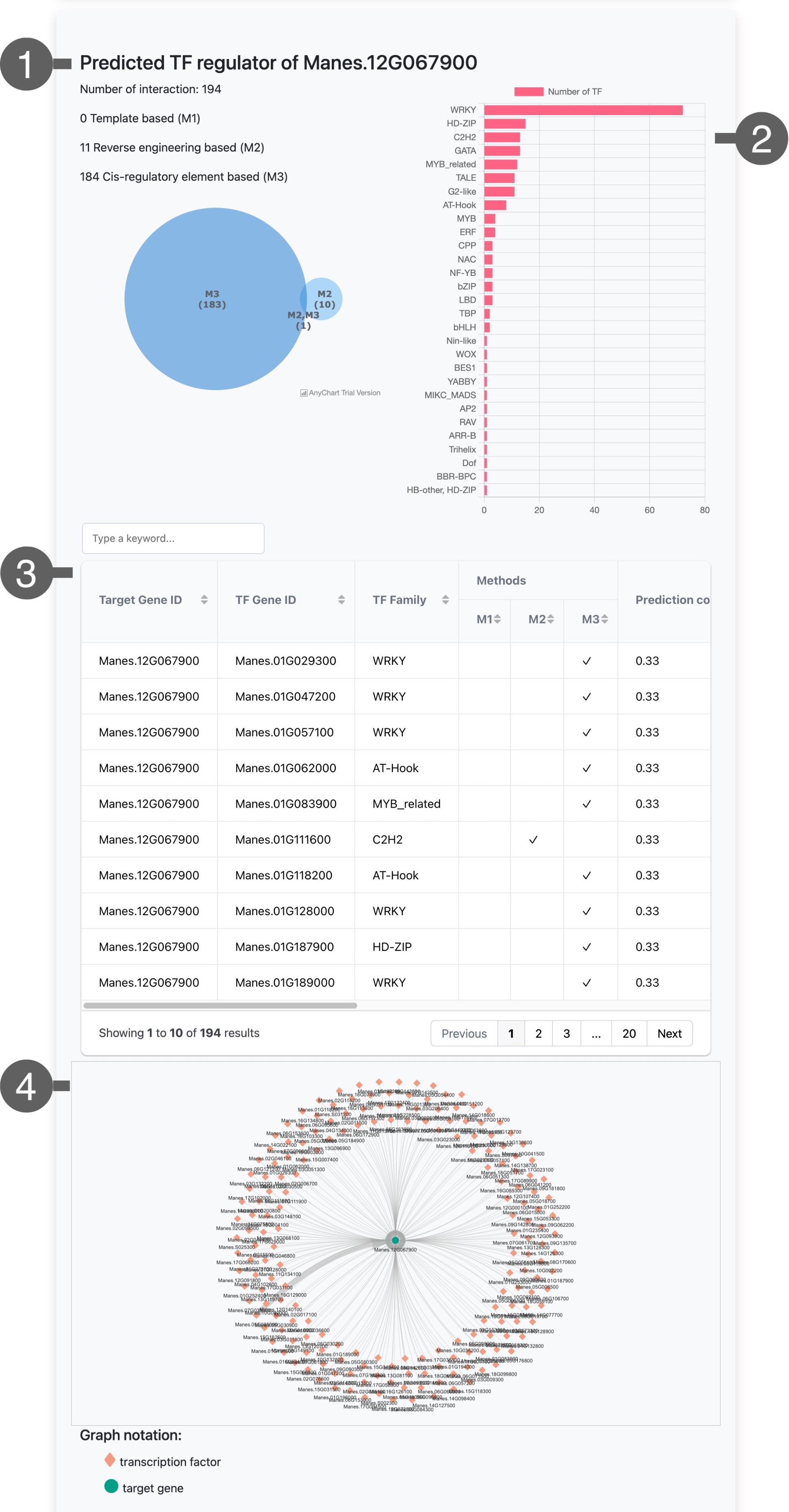
TRN Interaction information:
This session shows statistics of predicted MeTRN interaction of your input genes, including
- Statistic information of predicted target gene and transcription factor interactions based on 3 prediction approaches, template-based (M1), reverse engineering-based (M2), and cis-regulatory element-based (M3) methods.
- On the right side shows bar graph illustrate statistic information of predicted TF regulator genes classified by TF family, you can click and look only family of interest, the specific TF regulator genes of interested TF family will be shown in the below table.
- The interaction table included information of
- TF gene ID
- TF family
- Predicted methods (M1, M2, and M3)
- Consistency score based on three different methods
- Potential of gene expressed in 11 cassava tissues, including, leaf, shoot apical meristem (SAM), midvein, petiole, stem, lateral bud, storage root (SR), fibrous root (FR), root apical meristem (RAM), friable embryogenic callus (FEC), and organized embryogenic structures (OES) based on Wilson et al., 2013 dataset.
- Interaction in network view
- Blue-green circle node represent target gene
- Salmon Diamon represent transcription factor gene
For TRN regulated in pathway the table is shown
- Enzymatic gene information
- Gene ID
- Enzyme name
- EC number
- Their transcriptional regulator
- TF Gene ID
- TF family
- Statistic information of predicted target gene and transcription factor interactions based on 3 prediction approaches, template-based (M1), reverse engineering-based (M2), and cis-regulatory element-based (M3) methods
- Consistency score:
- 1.00 means predicted interaction from 3 methods
- 0.66 means predicted interaction from 2 methods
- 0.33 means predicted interaction from only one method
- Potential of gene expressed in 11 cassava tissues, including, leaf, shoot apical meristem (SAM), midvein, petiole, stem, lateral bud, storage root (SR), fibrous root (FR), root apical meristem (RAM), friable embryogenic callus (FEC), and organized embryogenic structures (OES) based on Wilson et al., 2013 dataset.
Description of terminology
Target Gene ID
TF Gene ID
M1
M2
M3
Found in tissue
Consistency score
\(M_i \) represents binary representation 1 if can predict by method \(i \). \(i \) represents 3 prediction approaches, template-based (\(M_1 \)), reverse engineering-based (\(M_2 \)), and cis-regulatory element-based (\(M_3 \)) methods that possible in interaction prediction.